4.1: Forces of Evolution
- Last updated
- Save as PDF
- Page ID
- 136392
Andrea J. Alveshere, Ph.D., Western Illinois University
Learning Objectives
- Define populations and population genetics as well as the methods used to study them.
- Identify the forces of evolution and become familiar with examples of each and their evolutionary significance.
- Explain how allele frequencies can be used to study evolution as it happens.
- Contrast micro- and macroevolution.
It’s hard for us, with our typical human life spans of less than 100 years, to imagine all the way back, 3.8 billion years ago, to the origins of life. Scientists still study and debate how life came into being and whether it originated on Earth or in some other region of the universe (including some scientists who believe that studying evolution can reveal the complex processes that were set in motion by God or a higher power). What we do know is that a living single-celled organism was present on Earth during the early stages of our planet’s existence. This organism had the potential to reproduce by making copies of itself, just like bacteria, many amoebae, and our own living cells today. In fact, with today’s genetic and genomic technologies, we can now trace genetic lineages, or phylogenies, and determine the relationships between all of today’s living organisms—eukaryotes (animals, plants, fungi, etc.), archaea, and bacteria—on the branches of the phylogenetic tree of life (Figure 4.1).
Looking at the common sequences in modern genomes, we can even make educated guesses about what the genetic sequence of the first organism, or universal ancestor of all living things, would likely have been. Through a wondrous series of mechanisms and events, that first single-celled organism gave rise to the rich diversity of species that fill the lands, seas, and skies of our planet. This chapter explores the mechanisms by which that amazing transformation occurred and considers some of the crucial scientific experiments that shaped our current understanding of the evolutionary process.
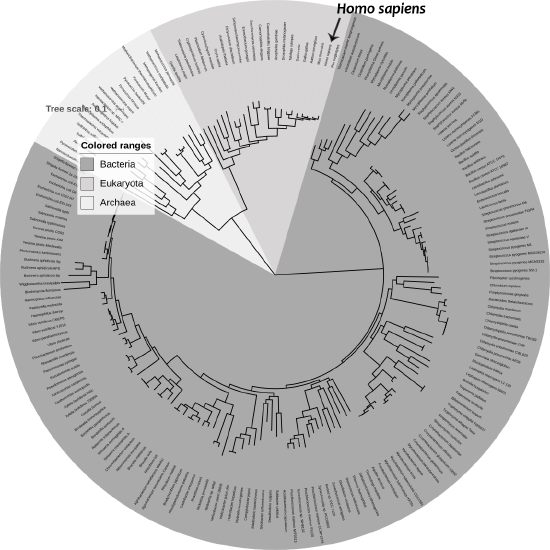 Figure \(\PageIndex{1}\): Phylogenetic tree of life.
Figure \(\PageIndex{1}\): Phylogenetic tree of life.About the Author
Andrea Alveshere
Western Illinois University, a-alveshere@wiu.edu, http://www.wiu.edu/cas/anthropology/faculty-staff/alveshere.php

Dr. Andrea Alveshere is an assistant professor of anthropology and chemistry at Western Illinois University. Her research focuses on relationships between humans and their environments, including questions of diet and health; cultural and biological adaptations; genetic disorders such as Neurofibromatosis Type 1 (NF1); effects of environmental factors on the preservation of bones, plant remains, and the molecules within them; and the comparative utility of various field and laboratory techniques to produce informative archaeological, nutritional, and forensic data.
Dr. Alveshere earned her B.A. in anthropology at the University of Washington with an emphasis in archaeology and an undergraduate research focus on the analysis of skeletal remains and geoarchaeological deposits. At the University of Minnesota, she completed her Ph.D. in anthropology, with a minor in human genetics. Her graduate thesis investigated factors that influence the preservation and detection of DNA in ancient and forensic specimens.
In addition to her academic experience, Dr. Alveshere worked for several years as a forensic scientist in the DNA/Biology section of the Minnesota Bureau of Criminal Apprehension Forensic Science Laboratory. She leads the WIU Archaeological Field School, which is offered every other summer, and has also conducted archaeological excavations in Israel, South Africa, and throughout the Midwestern United States.
For Further Exploration
Explore Evolution. HHMI’s Biointeractive. https://www.hhmi.org/biointeractive/evolution-collection
Teaching Evolution Through Human Examples. Smithsonian Museum of Natural History. http://humanorigins.si.edu/education/teaching-evolution-through-human-examples
References
Bloomfield, Gareth, David Traynor, Sophia P. Sander, Douwe M. Veltman, Justin A. Pachebat, and Robert R. Kay. 2015. “Neurofibromin Controls Macropinocytosis and Phagocytosis in Dictyostelium.” eLife 4:e04940.
Castle, W. E., and J. C. Phillips. 1914. Piebald Rats and Selection: An Experimental Test of the Effectiveness of Selection and of the Theory of Gametic Purity in Mendelian Crosses. Carnegie Institute of Washington, No. 195. Washington, DC: Carnegie Institute of Washington.
Chaix, Raphaëlle, Chen Cao, and Peter Donnelly. 2008. “Is Mate Choice in Humans MHC-Dependent?” PLoS Genetics 4 (9): e1000184.
Cook, Laurence M. 2003. “The Rise and Fall of the Carbonaria Form of the Peppered Moth.” The Quarterly Review of Biology 78 (4): 399–417.
Cota, Bruno Cézar Lage, João Gabriel Marques Fonseca, Luiz Oswaldo Carneiro Rodrigues, Nilton Alves de Rezende, Pollyanna Barros Batista, Vincent Michael Riccardi, Luciana Macedo de Resende. 2018. “Amusia and Its Electrophysiological Correlates in Neurofibromatosis Type 1.” Arquivos de Neuro-Psiquiatria 76 (5): 287–295.
Darwin, Charles. 1859. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray.
D’Asdia, Maria Cecilia, Isabella Torrente, Federica Consoli, Rosangela Ferese, Monia Magliozzi, Laura Bernardini, Valentina Guida, et al. 2013. “Novel and Recurrent EVC and EVC2 Mutations in Ellis-van Creveld Syndrome and Weyers Acrofacial Dyostosis.” European Journal of Medical Genetics 56 (2): 80–87.
Dobzhansky, Theodosius (1937). Genetics and the Origin of Species. Columbia University Biological Series. New York: Columbia University Press.
Facon, Benoît, Laurent Crespin, Anne Loiseau, Eric Lombaert, Alexandra Magro, Arnaud Estoup. 2011. “Can Things Get Worse When an Invasive Species Hybridizes? The Harlequin Ladybird Harmonia axyridis in France as a Case Study.” Evolutionary Applications 4 (1): 71–88.
Fisher, Ronald A. (1919). “The Correlation Between Relatives on the Supposition of Mendelian Inheritance.” Transactions of the Royal Society of Edinburgh 52 (2): 399–433.
Ford, E. B. 1942. Genetics for Medical Students. London: Methuen.
Ford, E. B. 1949. Mendelism and Evolution. London: Methuen.
Grant, Bruce S. 1999. “Fine-tuning the Peppered Moth Paradigm.” Evolution 53 (3): 980–984.
Haldane, J. B. S. 1924. “A Mathematical Theory of Natural and Artificial Selection (Part 1).” Transactions of the Cambridge Philosophical Society 23:19–41.
Huxley, Julian. 1942. Evolution: The Modern Synthesis. London: Allen & Unwin.
Imperato-McGinley, J., and Y.-S. Zhu. 2002. “Androgens and Male Physiology: The Syndrome of 5 Alpha-Reductase-2 Deficiency.” Molecular and Cellular Endocrinology 198: 51–59.
Jablonski, David, and W. G. Chaloner. 1994. “Extinctions in the Fossil Record [and Discussion].” Philosophical Transactions of the Royal Society of London B: Biological Sciences 344 (1,307): 11–17.
Livi-Bacci, Massimo. 2006. “The Depopulation of Hispanic America After the Conquest.” Population Development and Review 32 (2): 199–232.
Lombaert, Eric, Thomas Guillemaud, Jean-Marie Cornuet, Thibaut Malausa, Benoît Facon, and Arnaud Estoup. 2010. “Bridgehead Effect in the Worldwide Invasion of the Biocontrol Harlequin Ladybird.” PLoS ONE 5 (3): e9743.
Martins, Aline Stangherlin, Ann Kristine Jansen, Luiz Oswaldo Carneiro Rodrigues, Camila Maria Matos, Marcio Leandro Ribeiro Souza, Juliana Ferreira de Souza, Maria de Fátima Haueisen Sander Diniz, et al. 2016. “Lower Fasting Blood Glucose in Neurofibromatosis Type 1.” Endocrine Connections 5 (1): 28–33.
Morgan, T. H. 1911. “Random Segregation Versus Coupling in Mendelian Inheritance.” Science 34 (873): 384.
Pickering, Gary, James Lin, Roland Riesen, Andrew Reynolds, Ian Brindle, and George Soleas. 2004. “Influence of Harmonia axyridis on the Sensory Properties of White and Red Wine.” American Journal of Enology and Viticulture 55 (2): 153–159.
Repunte-Canonigo Vez, Melissa A. Herman, Tomoya Kawamura, Henry R. Kranzler, Richard Sherva, Joel Gelernter, Lindsay A. Farrer, Marisa Roberto, and Pietro Paolo Sanna. 2015. “NF1 Regulates Alcohol Dependence-Associated Excessive Drinking and Gamma-Aminobutyric Acid Release in the Central Amygdala in Mice and Is Associated with Alcohol Dependence in Humans.” Biological Psychiatry 77 (10): 870–879.
Riccardi, Vincent M. 1992. Neurofibromatosis: Phenotype, Natural History, and Pathogenesis. Baltimore: Johns Hopkins University Press.
Sanford, Malcolm T. 2006. “The Africanized Honey Bee in the Americas: A Biological Revolution With Human Cultural Implications, Part V—Conclusion.” American Bee Journal 146 (7): 597–599.
Sanna, Pietro Paolo, Cindy Simpson, Robert Lutjens, and George Koob. 2002. “ERK Regulation in Chronic Ethanol Exposure and Withdrawal.” Brain Research 948 (1–2): 186–191.
Weismann, August. 1892. Das Keimplasma: Eine Theorie der Vererbung [The Germ Plasm: A Theory of Inheritance]. Jena: Fischer.
World Health Organization. 1996. “Control of Hereditary Disorders: Report of WHO Scientific meeting (1996).” WHO Technical Reports 865. Geneva: World Health Organization.
World Health Organization. 2017. “Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics.” Global Priority Pathogens List, February 27. Geneva: World Health Organization. https://www.who.int/medicines/public...-ET_NM_WHO.pdf
Wright, Sewall. 1932. “The Roles of Mutation, Inbreeding, Crossbreeding, and Selection in Evolution.” Proceedings of the Sixth International Congress on Genetics 1 (6): 356–366.
Acknowledgment
Many thanks to Dr. Vincent M. Riccardi for sharing his vast knowledge of neurofibromatosis and for encouraging me to explore it from an anthropological perspective.
Figure Attributions
Figure 4.1 Tree of life SVG by Ivica Letunic: Iletunic, retraced by Mariana Ruiz Villarreal: LadyofHats, has been designated to the public domain (CC0). This item has been modified (made grayscale, rotated, labels added).
Figure 4.2A Lamarckian Evolution original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.2B Modern Synthesis original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.3 Weismann’s mouse-tail experiment original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.4 Castle’s Hooded Rat Experiment original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.5 Morgan’s Mutant Fruit Flies original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.6 UV-induced Thymine dimer mutation original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.7 Cytosine-to-thymine point mutation original to An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.8 Point and frameshift mutations original to An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.9 Woman with cutaneous neurofibromas (syptom of NF1) by Rick Guidotti of Positive Exposure is used by permission and available here is under a CC BY-NC 4.0 License.
Figure 4.10a Man with plexiform neurofibroma (syptom of NF1) is used by permission from Ashok Shrestha and available here is under a CC BY-NC 4.0 License.
Figure 4.10b Childhood photo of the same man with NF1 disorder is used by permission from Ashok Shrestha and available here is under a CC BY-NC 4.0 License.
Figure 4.11 Child with café-au-lait macules (birthmarks) typical of the earliest symptoms of NF1 by Andrea J. Alveshere is under a CC BY-NC 4.0 License.
Figure 4.12 The Cretaceous–Paleogene extinction event original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.13 6 Finger by Wilhelmy is under a CC BY-SA 4.0 License.
Figure 4.14 Ladybug Gene Flow original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.15 Peppered moths c2 by Khaydock is under a CC BY-SA 3.0 License.
Figure 4.16 Biology (ID: 185cbf87-c72e-48f5-b51e-f14f21b5eabd@9.17) by CNX OpenStax is used under a CC BY 4.0 License.
Figure 4.17 Sickle-cell smear 2015-09-10 by Paulo Henrique Orlandi Mourao, contrast modified and labels added by Katie Nelson, CC BY-SA 4.0.
Figure 4.18 Peacock tail advantage and disadvantages soriginal to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.19 Isolation Leading to Speciation original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.20 Darwin’s finches original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.
Figure 4.21 Ladybug mix original to Explorations: An Open Invitation to Biological Anthropology by Mary Nelson is under a CC BY-NC 4.0 License.

