3.6: End of Chapter of Review
- Last updated
- Save as PDF
- Page ID
- 158735
Learning Objectives
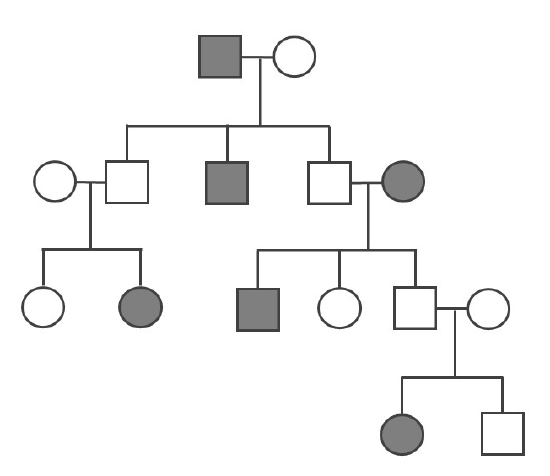
Figure 3.6.1
- What is the purpose of DNA replication? Explain in a few sentences what happens during DNA replication. When do DNA mutations happen? And how does this create phenotypic variation (i.e., different phenotypes of the same physical trait)?
- Using your own words, what are homologous chromosomes and sister chromatids? What are the key differences between mitosis and meiosis?
- Determine if the pedigree diagram below represents an autosomal dominant, autosomal recessive, or X-linked recessive pattern of inheritance. You should write the genotype (i.e., AA, Aa, or aa) above each square to help you (note: there may sometimes be two possible answers for a square’s genotype). Please also explain why you concluded that particular pattern of inheritance.
- Use base pairing rules to transcribe the following DNA template sequence into mRNA: GTAAAGGTGCTGGCCATC. Next, use the protein codon table (Figure 3.6.1) to translate the sequence. In regard to transcription, explain what the significance is of the first and last codon/protein in the sequence.
- In your opinion, what do you think the benefits are of direct-to-consumer (DTC) genetic testing? What are the drawbacks and/or greater ethical concerns? Do you think benefits outweigh concerns?
- Imagine that you submit your DNA sample to a genetic testing company and among the various diseases for which they test, there is an allele that is associated with late-onset Alzheimer’s disease. You have the option to view your Alzheimer’s result or to not view your result. What do you do and why?
GLOSSARY
Adenosine triphosphate (ATP): A high-energy compound produced by mitochondria that powers cellular processes.
Amino acids: Organic molecules that are the building blocks of protein. Each of the 20 different amino acids have their own unique chemical property. Amino acids are also chained together to form proteins.
Ancient (aDNA): DNA that is isolated from organic remains often dating from hundreds to thousands of years ago. Also, aDNA is typically degraded (i.e., damaged) due to exposure to the elements such as heat, acidity, and humidity.
Aneuploid: A cell with an unexpected amount of chromosomes. The loss or gain of chromosomes can occur during mitotic or meiotic division.
Antibodies: Immune-related proteins that can detect and bind to foreign substances in the blood such as pathogens.
Apoptosis: A series of molecular steps that is activated leading to cell death. Apoptosis can be activated when a cell fails checkpoints during the cell cycle; however, cancer cells have the ability to avoid apoptosis.
Autosomal: Refers to a pattern of inheritance where an allele is located on an autosome.
Autosomes: The numbered chromosomes, as opposed to the sex chromosomes.
Base pairs: Chemical bonding between nucleotides, like adenine (A) and thymine (T) or cytosine (C) and guanine (G) in DNA; or (A) and uracil (U) in RNA
Carbohydrate: Molecules composed of carbon and hydrogen atoms that can be broken down to supply energy.
Carrier: An individual who has a heterozygous genotype that is typically associated with a disease.
Cell cycle: A cycle the cell undergoes with checkpoints between phases to ensure that DNA replication and cell division occur properly.
Cell surface antigen: A protein that is found on a red blood cell’s surface.
Centromere: A structural feature that is defined as the “center” of a chromosome and which creates two different arm lengths. Term also refers to the region of attachment for microtubules during mitosis and meiosis.
Chromatin: DNA wrapped around histone complexes. During cell division, chromatin becomes a condensed chromosome.
Chromosome: DNA molecule that is wrapped around protein complexes, including histones.
Codominance: The effects of both alleles in a genotype can be seen in the phenotype.
Codons: A sequence that comprises three DNA nucleotides that together code for a protein; provide encoding instructions for the addition of one amino acid to a protein or indicating that the protein is complete.
Complex diseases: A category of diseases that are polygenic and are also influenced by environment and lifestyle factors.
Cytoplasm: The “jelly-like” matrix inside of the cell that contains many organelles and other cellular molecules.
Deleterious: A mutation that increases an organism’s susceptibility to disease.
Deoxyribonucleic acid (DNA): A molecule that carries the hereditary information passed down from parents to offspring. DNA can be described as a “double helix”’ shape. It includes two chains of nucleotides held together by hydrogen bonds with a sugar-phosphate backbone.
Diploid: Refers to an organism or cell with two sets of chromosomes.
DNA methylation: Methyl groups bind DNA, which modifies the transcriptional activity of a gene by turning it “on” or “off.”
DNA polymerase: Enzyme that adds nucleotides to existing nucleic acid strands during DNA replication. These enzymes can be distinguished by their processivity (e.g., DNA replication).
DNA replication: Cellular process in which DNA is copied and doubled.
DNA sequence: The order of nucleotide bases. A DNA sequence can be short, long, or representative of entire chromosomes or organismal genomes.
Dominant: Refers to an allele for which one copy is sufficient to be visible in the phenotype.
Elongation: The assembly of new DNA from template strands with the help of DNA polymerases.
Enzymes: Proteins responsible for catalyzing (accelerating) various biochemical reactions in cells.
Epigenetic profile: The methylation pattern throughout a genome—that is, which genes (and other genomic sites) are methylated and unmethylated.
Euchromatin: Loosely coiled chromosomes found within the nucleus that is accessible for regulatory processing of DNA.
Eukaryote: Single-celled or multicelled organism characterized by a distinct nucleus, with each organelle surrounded by its own membrane.
Exon: The DNA sequences within a gene that directly encode protein sequences. After being transcribed into messenger RNA, the introns are clipped out, and the exons are pasted together prior to translation.
Gametes: Haploid cells referred to as an egg and sperm that will fuse together during sexual reproduction to form a diploid organism.
Genetic recombination: A cellular process that occurs during meiosis I in which homologous chromosomes pair up and sister chromatids on different chromosomes physically swap genetic information.
Genome: All the genetic information of an organism.
Genotyping: A molecular procedure that is performed to test for the presence of certain alleles or to discover new ones.
Haploid: Cell or organism with one set of chromosomes (n = 23).
Helicase: A protein that breaks the hydrogen bonds that hold double-stranded DNA together.
Heterozygous: Genotype that consists of two different alleles.
Histones: Protein that DNA wraps around to assist with DNA organization within the nucleus.
Homologous chromosomes: A matching pair of chromosomes wherein one chromosome is maternally inherited and the other is paternally inherited.
Homozygous: Genotype that consists of two identical alleles.
Incomplete dominance: Heterozygous genotype that produces a phenotype that is a blend of both alleles.
Initiation: The recruitment of proteins to separate DNA strands and begin DNA replication.
Interphase: Preparatory period of the cell cycle when increased metabolic demand allows for DNA replication and doubling of the cell prior to cell division.
Introns: DNA sequences within a gene that do not directly encode protein sequences. After being transcribed into messenger RNA, the introns are clipped out, and the exons are pasted together prior to translation.
Karyotyping: The microscopic procedure where the number of chromosomes in a cell is determined.
Lagging strand: DNA template strand that is opposite to the leading strand. Therefore, DNA replication proceeds discontinuously, generating Okazaki fragments.
Leading strand: DNA template strand in which replication proceeds continuously.
Lipids: Fatty acid molecules that serves various purposes in the cell, including energy storage, cell signaling, and structure.
Meiosis: The process that gametes undergo to divide. The end of meiosis results in four haploid daughter cells.
Mendelian genetics: A classification given to phenotypic traits that are controlled by a single gene.
Messenger RNA (mRNA): RNA molecule that is transcribed from DNA. Its tri-nucleotide codons are “read” by a ribosome to build a protein.
Microarray technology: A genotyping procedure that utilizes a microarray chip, which is a collection of thousands of short nucleotide sequences attached to a solid surface that can probe genomic DNA.
Microbiome: The collective genomes of the community of microorganisms that humans have living inside of their body.
Mitochondrial DNA (mtDNA): Circular DNA segment found in mitochondria of a cell that is inherited maternally.
Mitochondrion: Specialized cellular organelle that is the site for energy production. It also has its own genome (mtDNA).
Mitosis: The process that somatic cells undergo to divide. The end of mitosis results in two diploid daughter cells.
Mutation: A nucleotide sequence variation from the template DNA strand that can occur during replication. Mutations can also happen during recombination.
Next-generation sequencing: A genotyping technology that involves producing millions of nucleotide sequences (from a single DNA sample) that are then read with a sequencing machine. It can be used for analyzing entire genomes or specific regions and requires extensive program-based applications.
Nuclear envelope: A double-layered membrane that encircles the nucleus.
Nucleic acid: A complex structure (like DNA or RNA) that carries genetic information about a living organism.
Nucleotide: The basic structural component of nucleic acids, which includes DNA (A, T, C, and G) and RNA (A, U, C, and G).
Nucleus: Double-membrane cellular organelle that helps protect DNA and regulation of nuclear activities.
Okazaki fragment: Short DNA strands derived from DNA replication on the lagging strand. They were discovered by Reiji and Tsuneko Okazaki in the 1960s.
Organelle: A structure within a cell that performs specialized tasks that are essential for the cell. There are different types of organelles with their own function.
Pathogenic: A genetic mutation (i.e., allele) that has a harmful phenotypic disease-causing effect.
Penetrance: The proportion of how often the possession of an allele results in an expected phenotype. Some alleles are more penetrant than others.
Phospholipid bilayer: Two layers of lipids that form a barrier due to the properties of a hydrophilic (water-loving) head and a hydrophobic (water-repelling) tail.
Polygenic trait: A phenotype that is controlled by two or more genes.
Polymerase chain reaction (PCR): A molecular biology procedure that can make copies of genomic DNA segments. A small amount of DNA is used as a starting template and is then used to make millions of copies.
Primer: A primer is a small sequence of nucleotides that bind DNA to start the process of DNA replication or PCR.
Prokaryote: A single-celled organism characterized by lack of a nucleus and membrane-enclosed organelles.
Promoter: The region of a gene that initiates transcription. Transcription factors can bind and DNA methylation may occur at a promoter site, which can modify the transcriptional activities of a gene.
Protein: Chain of amino acids that fold into a three dimensional structure that allow a cell to function in a variety of ways.
Protein synthesis: A multi-step process by which amino acids are strung together by RNA machinery read from a DNA template.
Recessive: Refers to an allele whose effect is not normally seen unless two copies are present in an individual’s genotype.
Ribonucleic acid (RNA): Single-stranded nucleic acid molecule.There are different RNAs found within cells and they perform a variety of functions, such as cell signaling and involvement in protein synthesis.
Ribosomal RNA (rRNA): A ribosome-bound molecule that is used to correctly assemble amino acids into proteins.
Ribosome: An organelle in the cell found in the cytoplasm or endoplasmic reticulum. It is responsible for reading mRNA and protein assemblage.
RNA polymerase: An enzyme that catalyzes the process of making RNA from a DNA template.
Sanger-sequencing: A process that involves the usage of fluorescently labeled nucleotides to visualize DNA (PCR fragments) at the nucleotide level.
Semi-conservative replication: DNA replication in which new DNA is replicated from an existing DNA template strand.
Sequencing: A molecular laboratory procedure that produces the order of nucleotide bases (i.e., sequences).
Sister chromatids: During DNA replication, sister chromatids are produced on the chromosome. In cell division, sister chromatids are pulled apart so that two cells can be formed. In meiosis, sister chromatids are also the sites of genetic recombination.
Somatic cells: Diploid cells that comprise body tissues and undergo mitosis for maintenance and repair of tissues.
Splicing: The process by which mature mRNAs are produced. Introns are removed (spliced) and exons are joined together.
Sugar phosphate backbone: A biochemical structural component of DNA. The “backbone” consists of deoxyribose sugars and phosphate molecules.
Telomere: A compound structure located at the ends of chromosomes to help protect the chromosomes from degradation after every round of cell division.
Termination: The halt of DNA replication activity that occurs when a DNA sequence “stop” codon is encountered.
Tissue: A cluster of cells that are morphologically similar and perform the same task.
Transcription: The process by which DNA nucleotides (within a gene) are copied, which results in a messenger RNA molecule.
Transcription factors: Proteins that bind to regulatory regions of genes (e.g., promoter) and increase or decrease the amount of transcriptional activity of a gene, including turning them “on” or “off.”
Transfer RNA (tRNA): RNA molecule involved in translation. Transfer RNA transports amino acids from the cell’s cytoplasm to a ribosome.
Translation: The process by which messenger RNA codons are read and amino acids are “chained together” to form proteins.
X-linked: Refers to a pattern of inheritance where the allele is located on the X or Y chromosome.
FOR FURTHER EXPLORATION
Websites
National Human Genome Research Institute https://www.genome.gov/
Genetics Home Reference https://ghr.nlm.nih.gov/
Genetics Generation http://knowgenetics.org/
yourgenome https://www.yourgenome.org/
Cardiovascular Disease: Genes are Important, but Health-Related Behaviors and Lifestyle Choices Can Make or Break Your Health http://ehrweb.aaas.org/ehr/books/3_howard.html
Gene Sequencing Speeds Diagnosis of Deadly Newborn Diseases http://www.pbs.org/wgbh/nova/next/body/newborn-gene-sequencing/
Carl Zimmer’s Game of Genomes https://www.statnews.com/feature/game-of-genomes/season-one/
Illumina Sequencing by Synthesis https://www.youtube.com/watch?v=fCd6B5HRaZ8
Articles
Aartsma-Rus, Annemieke, Ieke B. Ginjaar, and Kate Bushby. 2016. “The Importance of Genetic Diagnosis for Duchenne Muscular Dystrophy.” Journal of Medical Genetics 53 (3): 145–151.
Acuna-Hidalgo, Rocio, Joris A. Veltman, and Alexander Hoischen. 2016. “New Insights into the Generation and Role of De Novo Mutations in Health and Disease.” Genome Biology 17 (241): 1-19.
Albert, Benjamin, Susanna Tomassetti, Yvonne Gloor, Daniel Dilg, Stefano Mattarocci, Slawomir Kubik, Lukas Hafner, and David Shore. 2019. “Sfp1 Regulates Transcriptional Networks Driving Cell Growth and Division through Multiple Promoter-Binding Modes.” Genes & Development 33 (5–6): 288–293.
Almathen, Faisal, Haitham Elbir, Hussain Bahbahani, Joram Mwacharo, and Olivier Hanotte. 2018. “Polymorphisms in Mc1r and Asip Genes Are Associated With Coat Color Variation in the Arabian Camel.” Journal of Heredity 109 (6): 700–706.
Ballester, Leomar Y., Rajyalakshmi Luthra, Rashmi Kanagal-Shamanna, and Rajesh R. Singh. 2016. “Advances in Clinical Next-Generation Sequencing: Target Enrichment and Sequencing Technologies.” Expert Review of Molecular Diagnostics 16 (3): 357–372.
Baranovskiy, Andrey G., Vincent N. Duong, Nigar D. Babayeva, Yinbo Zhang, Youri I. Pavlov, Karen S. Anderson, and Tahir H. Tahirov. 2018. “Activity and Fidelity of Human DNA Polymerase Alpha Depend on Primer Structure.” Journal of Biological Chemistry 293 (18): 6,824–6,843.
Brezina, Paulina R., Raymond Anchan, and William G. Kearns. 2016. “Preimplantation Genetic Testing for Aneuploidy: What Technology Should You Use and What Are the Differences?” Journal of Assisted Reproduction and Genetics 33 (7): 823–832.
Bultman, Scott J. 2017. “Interplay Between Diet, Gut Microbiota, Epigenetic Events, and Colorectal Cancer.” Molecular Nutrition & Food Research 61 (1):1-12.
Cutting, Garry R. 2015. “Cystic Fibrosis Genetics: From Molecular Understanding to Clinical Application.” Nature Reviews Genetics 16 (1): 45–56.
D’Alessandro, Giuseppina., and Fabrizio d’Adda di Fagagna. 2017. “Transcription and DNA Damage: Holding Hands or Crossing Swords?” Journal of Molecular Biology 429 (21): 3,215–3,229.
De Craene, Johan-Owen, Dimitri L. Bertazzi, Séverine Bar, and Sylvie Friant. 2017. “Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways.” International Journal of Molecular Sciences 18 (3): 1-20.
Deng, Lian, and Shuhua Xu. 2018. “Adaptation of Human Skin Color in Various Populations.” Hereditas 155 (1): 1-12.
Dever, Thomas E., Terri G. Kinzy, and Graham D. Pavitt. 2016. “Mechanism and Regulation of Protein Synthesis in Saccharomyces Cerevisiae.” Genetics 203 (1): 65–107.
Eme, Laura, Anja Spang, Jonathan Lombard, Courtney W. Stairs, and Thijs J. G. Ettema. 2017. “Archaea and the Origin of Eukaryotes.” Nature Reviews Microbiology 15 (12): 711–723.
Gomez-Carballa, Alberto, Jacobo Pardo-Seco, Stefania Brandini, Alessandro Achilli, Ugo A. Perego, Michael D. Coble, Toni M. Diegoli, et al. 2018. “The Peopling of South America and the Trans-Andean Gene Flow of the First Settlers.” Genome Research 28 (6): 767–779.
Gvozdenov, Zlata, Janhavi Kolhe, and Brian C. Freeman. 2019. “The Nuclear and DNA-Associated Molecular Chaperone Network.” Cold Spring Harbor Perspectives in Biology. New York:Cold Spring Harbor Laboratory Press.
Harkins, Kelly M., and Anne C. Stone. 2015. “Ancient Pathogen Genomics: Insights Into Timing and Adaptation.” Journal of Human Evolution 79: 137–149.
Jackson, Maria, Leah Marks, Gerhard H. W. May, and Joanna B. Wilson. 2018. “The Genetic Basis of Disease.” Essays in Biochemistry 62 (5): 643–723.
Lenormand, Thomas., Jan Engelstadter, Susan E. Johnston, Erik Wijnker, and Christopher R. Haag. 2016. “Evolutionary Mysteries in Meiosis.” Philosophical Transactions of the Royal Society B 371: 1-14.
Levy, Shawn E., and Richard M. Myers. 2016. “Advancements in Next-Generation Sequencing.” Annual Review of Genomics and Human Genetics 17: 95–115.
Lu, Mengfei, Cathryn M. Lewis, and Matthew Traylor. 2017. “Pharmacogenetic Testing Through the Direct-to-Consumer Genetic Testing Company 23andme.” BMC Medical Genomics 10 (47): 1-8.
Ly, Lundi, Donovan Chan, Mahmoud Aarabi, Mylene Landry, Nathalie A. Behan, Amanda J. MacFarlane, and Jacquetta Trasler. 2017. “Intergenerational Impact of Paternal Lifetime Exposures to Both Folic Acid Deficiency and Supplementation on Reproductive Outcomes and Imprinted Gene Methylation.” Molecular Human Reproduction 23 (7): 461–477.
Ma, Wenxiu, Giancarlo Bonora, Joel B. Berletch, Xinxian Deng, William S. Noble, and Christine M. Disteche. 2018. “X-Chromosome Inactivation and Escape From X Inactivation in Mouse.” Methods in Molecular Biology 1,861: 205–219.
Machiela, Mitchell J., Weiyin Zhou, Eric Karlins, Joshua N. Sampson, Neal D. Freedman, Qi Yang, Belynda Hicks, et al. 2016. “Female Chromosome X Mosaicism Is Age-Related and Preferentially Affects the Inactivated X Chromosome.” Nat Commun 7: 1-9. doi: 10.1038/ncomms11843.
Mahdavi, Morteza, Mohammadreza Nassiri, Mohammad M. Kooshyar, Masoume Vakili-Azghandi, Amir Avan, Ryan Sandry, Suja Pillai, Alfred K. Lam, and Vinod Gopalan. 2019. “Hereditary Breast Cancer; Genetic Penetrance and Current Status With BRCA.” Journal of Cellular Physiology 234 (5): 5,741–5,750.
McDade, Thomas W., Calen P. Ryan, Meaghan J. Jones, Morgan K. Hoke, Judith Borja, Gregory E. Miller, Christopher W. Kuzawa, and Michael S. Kobor. 2019. “Genome-Wide Analysis of DNA Methylation in Relation to Socioeconomic Status During Development and Early Adulthood.” American Journal of Physical Anthropology 169 (1): 3–11.
Migeon, Barbara R. 2017. “Choosing the Active X: The Human Version of X Inactivation.” Trends in Genetics 33 (12): 899–909.
Myerowitz, Rachel. 1997. “Tay-Sachs Disease-Causing Mutations and Neutral Polymorphisms in the Hex a Gene.” Human Mutation 9: 195–208.
Onufriev, Alexey V. and Helmut Schiessel. 2019. “The Nucleosome: From Structure to Function Through Physics.” Current Opinion in Structural Biology 56: 119–130.
Quillen, Ellen E., Heather L. Norton, Esteban J. Parra, Frida Lona-Durazo, Khai C. Ang, Florin M. Illiescu, Laurel N. Pearson, et al. 2019. “Shades of Complexity: New Perspectives on the Evolution and Genetic Architecture of Human Skin.” American Journal of Physical Anthropology 168 (67): 4–26.
Raspelli, Erica and Roberta Fraschini. 2019. “Spindle Pole Power in Health and Disease.” Current Genetics 65 (4): 851-855.
Ravinet, M., R. Faria, R. K. Butlin, J. Galindo, N. Bierne, M. Rafajlovic, M. A. F. Noor, B. Mehlig, and A. M. Westram. 2017. “Interpreting the Genomic Landscape of Speciation: A Road Map for Finding Barriers to Gene Flow.” Journal of Evolutionary Biology 30 (8): 1,450–1,477.
Regev, Aviv, Sarah A. Teichmann, Eric S. Lander, Ido Amit, Christophe Benoist, Ewan Birney, Bernd Bodenmiller, et al. 2017. “The Human Cell Atlas.” Elife 6e27041: 1-30. doi: doi.org/10.7554.eLife.27041.
Roberts, Andrea L., Nicole Gladish, Evan Gatev, Meaghan J. Jones, Ying Chen, Julia L. MacIsaac, Shelley S. Tworoger, et al. 2018. “Exposure to Childhood Abuse Is Associated With Human Sperm DNA Methylation.” Translational Psychiatry 8 (194): 1-11.
Roger, Andrew J., Sergio A. Muñoz-Gómez, and Ryoma Kamikawa. 2017. “The Origin and Diversification of Mitochondria.” Current Biology 27 (21): R1177–R1192.
Ségurel, Laure and Céline Bon. 2017. “On the Evolution of Lactase Persistence in Humans.” Annual Review of Genomics and Human Genetics 18: 297–319.
Sheth, Bhavisha P. and Vrinda S. Thaker. 2017. “DNA Barcoding and Traditional Taxonomy: An Integrated Approach for Biodiversity Conservation.” Genome 60 (7): 618–628.
Snedeker, Jonathan, Matthew Wooten, and Xin Chen. 2017. “The Inherent Asymmetry of DNA Replication.” Annual Review of Cell and Developmental Biology 33: 291–318.
Sullivan-Pyke, Chantae and Anuja Dokras. 2018. “Preimplantation Genetic Screening and Preimplantation Genetic Diagnosis.” Obstetrics and Gynecology Clinics of North America 45 (1): 113–125.
Szostak, Jack W. 2017. “The Narrow Road to the Deep Past: In Search of the Chemistry of the Origin of Life.” Angewandte Chemie International Edition 56 (37): 11,037–11,043.
Tessema, Mathewos, Ulrich Lehmann, and Hans Kreipe. 2004. “Cell Cycle and No End.” Virchows Archiv European Journal of Pathology 444 (4): 313–323.
Tishkoff, Sarah A., Floyd A. Reed, Alessia Ranciaro, Benjamin F. Voight, Courtney C. Babbitt, Jesse S. Silverman, Kweli Powell, et al. 2007. “Convergent Adaptation of Human Lactase Persistence in Africa and Europe.” Nature Genetics 39 (1): 31–40.
Visootsak, Jeannie and John M. Graham, Jr. 2006. “Klinefelter Syndrome and Other Sex Chromosomal Aneuploidies.” Orphanet Journal of Rare Diseases 1:42 doi: 10.1186/1750-1172-1-42.
Yamamoto, Fumi-ichiro, Henrik Clausen, Thayer White, John Marken, and Sen-itiroh Hakomori. 1990. “Molecular Genetic Basis of the Histo-Blood Group Abo System.” Nature 345: 229–233.
Zlotogora, Joël. 2003. “Penetrance and Expressivity in the Molecular Age.” Genetics in Medicine 5: 347–352.
Zorina-Lichtenwalter, Katerina, Ryan N. Lichtenwalter, Dima V. Zaykin, Marc Parisien, Simon Gravel, Andrey Bortsov, and Luda Diatchenko. 2019. “A Study in Scarlet: MC1R as the Main Predictor of Red Hair and Exemplar of the Flip-Flop Effect.” Human Molecular Genetics 28 (12): 2,093-2,106.
Zwart, Haeh. 2018. “In the Beginning Was the Genome: Genomics and the Bi-Textuality of Human Existence.” New Bioethics 24 (1): 26–43.

