4.1.3: The Force of Evolution
- Last updated
- Save as PDF
- Page ID
- 136395
Today, we recognize that evolution takes place through a combination of mechanisms: mutation, genetic drift, gene flow, and natural selection. These mechanisms are called the “forces of evolution” and together they can account for all the genotypic variation observed in the world today. Keep in mind that each of these forces was first defined and then tested—and re-tested—through the experimental work of the many scientists who contributed to the Modern Synthesis.
Mutation
The first force of evolution we will discuss is mutation, and for good reason: Mutation is the original source of all the genetic variation found in every living thing. Let’s try again to imagine all the way back in time to the very first single-celled organism, floating in Earth’s primordial sea. Based on what we observe in simple, single-celled organisms today, that organism probably spent its lifetime absorbing nutrients and dividing to produce cloned copies of itself. While the numbers of individuals in that population would have grown (as long as the environment was favorable), nothing would have changed in that perfectly cloned population. There would not have been variety among the individuals. It was only through a copying error—the introduction of a mutation, or change, into the genetic code—that each new allele was introduced into the population.
When we think of genetic mutation, we often first think of deleterious mutations—the ones associated with negative effects such as the beginnings of cancers or heritable disorders. The fact is, though, that every genetic adaptation that has helped our ancestors survive since the dawn of life is directly due to a beneficial mutation—a changes in the DNA that provided some sort of advantage to a given population at a particular moment in time. For example, a beneficial mutation allowed chihuahuas and other tropical-adapted dog breeds to have much thinner fur coats than their cold-adapted cousins the northern wolves, malamutes, and huskies.
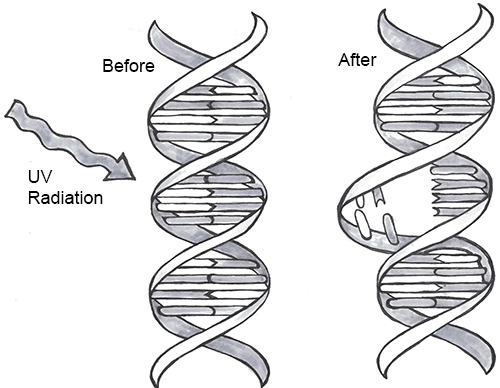
Every one of us has genetic mutations. Yes, even you. The DNA in some of your cells today differs from the original DNA that you inherited when you were a tiny, fertilized egg. Mutations occur all the time in the cells of our skin and other organs, due to chemical changes in the nucleotides. Exposure to the UV radiation in sunlight is one common cause of skin mutations. Interaction with UV light causes UV crosslinking, in which adjacent thymine bases bind with one another (Figure 4.6). Many of these mutations are detected and corrected by DNA repair mechanisms, enzymes that patrol and repair DNA in living cells, while other mutations may cause a new freckle or mole or, perhaps, an unusual hair to grow. For people with the autosomal recessive disease xeroderma pigmentosum, these repair mechanisms do not function correctly, resulting in a host of problems, especially related to sun exposure, including severe sunburns, dry skin, heavy freckling, and other pigment changes.
Most of our mutations exist in somatic cells, which are the cells of our organs and other body tissues. Those will not be passed on to future generations and so will not affect the population over time. Only mutations that occur in the gametes, the reproductive cells (i.e., the sperm or egg cells), will be passed on to future generations. When a new mutation pops up at random in a family lineage, it is known as a spontaneous mutation. If the individual born with this spontaneous mutation passes it on to his offspring, those offspring receive an inherited mutation. Geneticists have identified many classes of mutations and the causes and effects of many of these.
Point Mutations
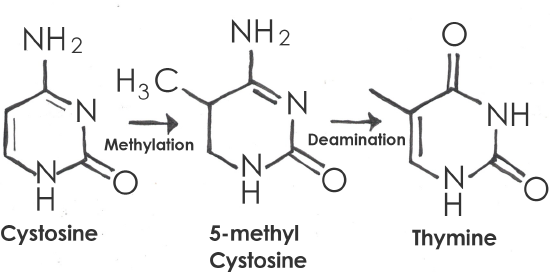
A point mutation is a single-letter (single-nucleotide) change in the genetic code resulting in the substitution of one nucleic acid base for a different one. As you learned in Chapter 3, the DNA code in each gene is translated through three-letter “words” known as codons. So depending on how the point mutation changes the codon, the effect it will have on the protein may be major or minor, or may make no difference at all. One of the most common causes of point mutations is a chemical change called cytosine methylation. In cytosine methylation, a methyl group is added to a cytosine base, which further converts to thymine after hydrolytic deamination (water-induced removal of an amine group; Figure 4.7). If this mutation is not detected before replication, half of the daughter cells will inherit a thymine (T) in the sequence where a cytosine (C) is usually located. This is one of the most common causes of the autosomal dominant disorder neurofibromatosis type 1 (NF1), discussed in Case Study #1 (see below).
If a mutation does not change the resulting protein, then it is called a synonymous mutation. Synonymous mutations do involve a letter (nucleic acid) change, but that change results in a codon that codes for the same “instruction” (the same amino acid or stop code) as the original codon. Mutations that do cause a change in the protein are known as . There are several classes of non-synonymous mutations, which are defined by their effects on the encoded protein: missense, nonsense, and splice site mutations (Figure 4.8).
A missense mutation produces a change in a single amino acid. In this case, the protein is assembled correctly, both before and after the point mutation, but one amino acid, encoded by the codon containing the point mutation, is incorrect. This may impact how the finished protein functions by, for example, preventing it from folding correctly and/or disrupting an enzyme binding site. Nonsense mutations convert codons that encode amino acids into stop codons, meaning that the protein will be assembled correctly up until the codon containing the mutation and then assembly will be prematurely terminated. Depending on where in the gene the nonsense mutation falls, this may have a major or very minor impact. A splice site mutation changes the genetic code so that the process of removing the intron sequences from the mRNA is disrupted. This can result in the erroneous inclusion of an intron sequence or the exclusion of one of the exons that should have been retained.
| Mutation Type | Illustration | Result |
| No mutation (normal DNA) | 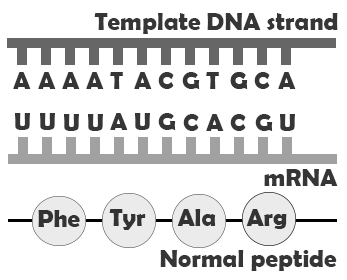 |
Normal protein produced |
| Synonymous (silent) mutation | 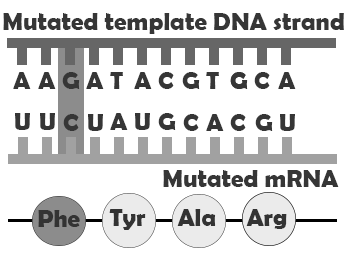 |
Normal protein produced |
| Missense mutation | 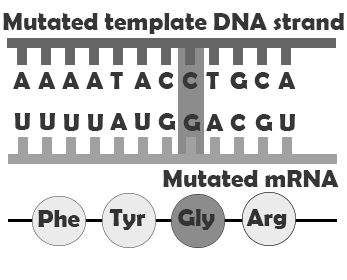 |
Slight difference in amino acid sequence |
| Nonsense mutation | 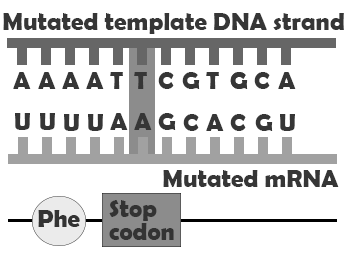 |
Protein terminates early |
| Mutation Type | Illustration | Result |
| Frameshift insertion | 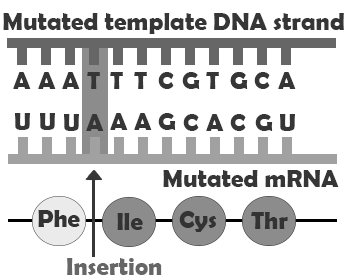 |
Major difference in amino acid sequence |
| Frameshift deletion | 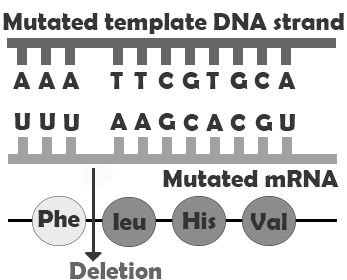 |
Major difference in amino acid sequence |
Insertions and Deletions
In addition to point mutations, another class of mutations are insertions and deletions, or indels, for short. As the name suggests, these involve the addition (insertion) or removal (deletion) of one or more coding sequence letters (nucleic acids). These typically first occur as an error in DNA replication, wherein one or more nucleotides are either duplicated or skipped in error.
Frameshift mutations are types of indels that involve the insertion or deletion of any number of nucleotides that is not a multiple of three. Because these indels are not consistent with the codon numbering, they “shift the reading frame,” causing all the codons beyond the mutation to be misread. These mutations can create extensive changes to the protein sequence, potentially not only causing it to lose function but also possibly creating new enzyme-binding sites, leading to new interactions between the protein and other components of the cellular environment. Like point mutations, small indels can also disrupt splice sites. Entire codons or sets of codons may also be removed or added if the indel is a multiple of three nucleotides.
Transposable Elements, or transposons, are fragments of DNA that can “jump” around in the genome. There are two types of transposons: Class I transposons, or retrotransposons, which are transcribed from DNA into RNA and then “reverse transcribed,” to insert the copied sequence into a new location in the DNA; and Class II transposons, or DNA transposons, which do not involve RNA— instead, DNA transposons are clipped out of the DNA sequence itself and inserted elsewhere in the genome. Because transposable elements insert themselves into (and, in the case of Class II transposons, remove themselves from) existing DNA sequences, they are frequent gene disruptors. At certain times, and in certain species, it appears that transposons became very active, likely accelerating the mutation rate (and thus, the genetic variation) in those populations during the active periods.
Chromosomal Alterations
The final major category of genetic mutations are changes at the chromosome level: crossover events, nondisjunction events, and translocations. Nondisjunction events occur when the homologous chromosomes (in meiosis I) or sister chromatids (in meiosis II and mitosis) fail to separate after pairing. The result is that both chromosomes or chromatids end up in the same daughter cell, leaving the other daughter cell without any copy of that chromosome. Most nondisjunctions at the gamete level are fatal to the embryo. The most widely known exception is Trisomy 21, or Down syndrome, which results from an embryo that inherits three copies of Chromosome 21: two from one parent (due to a nondisjunction event) and one from the other. Trisomies (triple chromosome conditions) of Chromosomes 18 (Edwards syndrome) and 13 (Patau syndrome) are also known to result in live births, but the children usually have severe complications and rarely survive beyond the first year of life. Sex chromosome trisomies (XXX, XXY, XYY) and X chromosome monosomies (inheritance of an X chromosome from one parent and no sex chromosome from the other) are also survivable and fairly common. The symptoms vary but often include atypical sexual characteristics, either at birth or at puberty, and often result in sterility. The X chromosome carries unique genes that are required for survival; therefore, Y chromosome monosomies are incompatible with life.
Chromosomal translocations involve transfers of DNA between non-homologous chromosomes. This may involve swapping large portions of two or more chromosomes. The exchanges of DNA may be balanced or unbalanced. In balanced translocations, the genes are swapped, but no genetic information is lost. In unbalanced translocations, there is an unequal exchange of genetic material resulting in duplication or loss of genes. Translocations result in new chromosomal structures called derivative chromosomes, because they are derived or created from two different chromosomes. Translocations are often found to be linked to cancers and can also cause infertility. Even if the translocations are balanced in the parent, the embryo often won’t survive unless the baby inherits both of that parent’s derivative chromosomes (to maintain the balance).
Case Study #1: Neurofibromatosis Type 1 (NF1)
, also known as , is a surprisingly common genetic disorder, affecting more people than cystic fibrosis and muscular dystrophy combined. Even more surprising, given how common it is, is how few people have heard of it. One in every 3,000 babies is born with NF1, and this holds true for all populations worldwide (Riccardi 1992). This means that, for every 3,000 people in your community, there is likely at least one community member living with this disorder. Approximately half of these cases are due to spontaneous mutations—that is, the person is the first in their family to have the disorder. The other half of the NF1 cases are inherited from a parent with this disorder. NF1 syndrome is an autosomal dominant condition, which means that everyone born with a mutation in the gene, whether inherited or spontaneous, has a 50:50 chance of passing the NF1 syndrome on to each of their children.
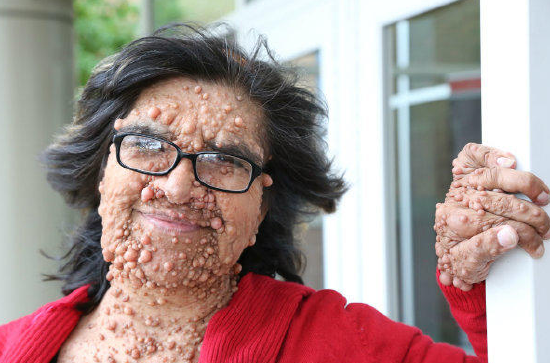 Figure \(\PageIndex{3}\): Photo of a woman with many cutaneous neurofibromas, a common symptom of Neurofibromatosis Type 1.
Figure \(\PageIndex{3}\): Photo of a woman with many cutaneous neurofibromas, a common symptom of Neurofibromatosis Type 1.The NF1 disorder results from disruption of the NF1 gene on Chromosome 17. Studies of individuals with NF1 have identified over 3,000 different mutations within the gene (including small and large indels, point mutations, and translocations). The NF1 gene is one of the largest known genes, containing at least 60 exons (protein-encoding sequences) in a span of about 300,000 nucleotides. It encodes a correspondingly large protein called neurofibromin. Neurofibromin is a fascinating protein, and we are still learning about all its functions.
Studying the symptoms in people who have mutations in an NF1 gene can provide important insights. There are two other types of Neurofibromatosis (Type 2 and Schwannomatosis) that involve some of the same symptoms but are much less common than NF1 and are not due to mutations in the same gene (or even the same chromosome).
 Figure \(\PageIndex{4}\): Photo of a man with large plexiform neurofibroma, another symptom of Neurofibromatosis Type 1.
Figure \(\PageIndex{4}\): Photo of a man with large plexiform neurofibroma, another symptom of Neurofibromatosis Type 1.We know that neurofibromin plays an important role in preventing tumor growth because, when a mutation occurs causing the NF1 disorder, one of the most common symptoms is the growth of benign (non-cancerous) tumors, called neurofibromas. Neurofibromas sprout from nerve sheaths—the tissues that encase our nerves—throughout the body. There is no way to predict where the tumors will occur, or when or how quickly they will grow, although only about 15% turn malignant (cancerous).
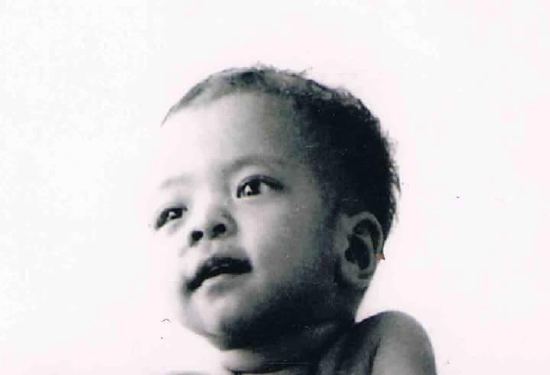
The two types of neurofibromas that are typically most visible are cutaneous neurofibromas, which are spherical bumps on, or just under, the surface of the skin (Figure 4.9), and plexiform neurofibromas, growths involving whole branches of nerves, often giving the appearance that the surface of the skin is “melting” (Figure 4.10).
Unfortunately, although research is ongoing, there is currently no cure for NF1. Surgical removal of neurofibromas risks paralysis, due to the high potential for nerve damage, and often results in the tumors growing back even more vigorously. This means that patients are often forced to live with disfiguring and often painful neurofibromas. People who are not familiar with NF1 often mistake neurofibromas for something contagious. This makes it especially hard for people living with NF1 to get jobs working with the public or even to enjoy spending time away from home. Raising public awareness about NF1 and its symptoms can be a great help in improving the quality of life for people living with this condition.

Babies who have NF1 rarely have neurofibromas, which often begin to grow during puberty. One of the first symptoms of NF1 in a small child is usually the appearance of , or CALS, which are flat, brown birthmark-like spots on the skin (Figure 4.11). CALS are often light brown, like the color of coffee with cream, which is the reason for the name, although the shade of the pigment depends on a person’s overall complexion. Some babies are born with CALS, but for others the spots appear within the first few years of life. Having six or more CALS larger than five millimeters (mm) across is a strong indicator that a child may have NF1 (the required size increases to 15 mm for diagnosis after puberty).
A second sign is always needed to confirm a clinical diagnosis of NF1. The second sign often comes in the form of freckles in unusual areas, such as the groin or underarms, or with the first appearance of neurofibromas. Other common symptoms include gliomas (tumors) of the optic nerve, which can cause vision loss; thinning of bones and failure to heal if they break (often requiring amputation); low muscle tone (poor muscle development, often delaying milestones such as sitting up, crawling, and walking); hearing loss, due to neurofibromas on auditory nerves; and learning disabilities, especially those involving spatial reasoning. Approximately 50 % of people with NF1 have some type of speech and/or learning disability and often benefit greatly from early intervention services. Intellectual disability, however, is not common with NF1, so most people with NF1 live independently as adults. Many people with NF1 live full and successful lives, as long as their symptoms can be managed.
Based on the wide variety of symptoms, it’s clear that neurofibromin plays important roles in many biochemical pathways. While everyone who has NF1 will exhibit some symptoms during their lifetime, there is a great deal of variation in the types and severity of symptoms, even between individuals from the same family who share the exact same NF1 mutation. It seems crazy that a gene with so many important functions would be so susceptible to mutation. Part of this undoubtedly has to do with its massive size—a gene with 300,000 nucleotides has ten times more nucleotides available for mutation than does a gene of 30,000 bases. This also suggests that the mutability of this gene might provide some benefits, which is a possibility that we will revisit later in this chapter.
Genetic Drift
The second force of evolution is commonly known as genetic drift. This is an unfortunate misnomer, as this force actually involves the drifting of alleles, not genes. Genetic drift refers to random changes (“drift”) in allele frequencies from one generation to the next. The genes are remaining constant within the population; it is only the alleles of the genes are changing in frequency. The random nature of genetic drift is a crucial point to understand: it specifically occurs when none of the variant alleles confer an advantage.
Let’s imagine far back in time, again, to that first population of living cells, subsisting and occasionally dividing, in the primordial sea. Many generations have passed, and mutations have created distinct chromosomes. The cells are now amoeba-like, larger than many of their tiny bacterial neighbors, who have long since become their favorite source of nutrients. A mutation occurs in one of the cells that changes the texture of the cell membrane from a relatively smooth surface to a highly ruffled one. This has absolutely no effect on the cell’s quality of life or ability to reproduce. In fact, eyes haven’t evolved yet, so no one in the world at the time would even notice the difference. The cells in the population continue to divide, and the offspring of the ruffled cell inherit the ruffled membrane. The frequency (%) of the ruffled allele in the population, from one generation to the next, will depend entirely on how many offspring that first ruffled cell ends up having, and the random events that might make the ruffled alleles more common or more rare (such as population bottlenecks and founder effects, discussed below).
Sexual Reproduction and Random Inheritance
Tracking alleles gets a bit more complicated in our primordial cells when, after a number of generations, a series of mutations have created populations that reproduce sexually. These cells go through an extra round of cell-division (meiosis) to create haploid gametes. The combination of two gametes, each containing half a set of homologous chromosomes, is required to produce each new diploid offspring. In the earlier population, which reproduced via asexual reproduction, a cell either carried the smooth allele or the ruffled allele. With sexual reproduction, a cell inherits one allele from each parent, so there are homozygous cells that contain two smooth alleles, homozygous cells that contain two ruffled alleles, and heterozygous cells that contain one of each allele. If the new, ruffled allele happens to be dominant (and we’ll imagine that it is), the heterozygotes will have ruffled cell phenotypes, but will have a 50:50 chance of passing on a smooth allele to each offspring.
In sexually reproducing populations (including humans and many other animals and plants in the world today), that 50:50 chance of inheriting one or the other allele from each parent plays a major role in the random nature of genetic drift.
Population Bottlenecks
A population bottleneck occurs when the number of individuals in a population drops dramatically due to some random event. The most obvious, familiar examples are natural disasters. Tsunamis and hurricanes devastating island and coastal populations and forest fires and river floods wiping out populations in other areas are all too familiar. When a large portion of a population is randomly wiped out, the allele frequencies (i.e., the percentages of each allele) in the small population of survivors are often much different from the frequencies in the pre-disaster, or “parent,” population. If such an event happened to our primordial ocean cell population—perhaps a volcanic fissure erupted in the ocean floor and only the cells that happened to be farthest from the spewing lava and boiling water survived—we might end up, by random chance, with a surviving population that had mostly ruffled alleles, in contrast to the parent population, which had only a small percentage of ruffles.

One of the most famous examples of a population bottleneck is the prehistoric disaster that led to the extinction of dinosaurs, the event (often abbreviated K–Pg; previously K-T). This occurred approximately 66 million years ago. Dinosaurs and all their neighbors were going about their ordinary routines when a massive asteroid zoomed in from space and crashed into what is now the Gulf of Mexico, creating an impact so enormous that populations within hundreds of miles of the crash site were likely immediately wiped out. The skies filled with dust and debris, causing temperatures to plummet worldwide. It’s estimated that 75% of the world’s species went extinct as a result of the impact and the deep freeze that followed (Jablonski and Chaloner 1994). The populations that emerged from the K-Pg extinction were markedly different from their pre-disaster communities. Surviving mammal populations expanded and diversified, and other new creatures appeared. The ecosystems of Earth filled with new organisms and have never been the same (Figure 4.12).
Much more recently in geological time, during the colonial period, many human populations experienced bottlenecks as a result of the fact that imperial powers were inclined to slaughter communities who were reluctant to give up their lands and resources. This effect was especially profound in the Americas, where indigenous populations faced the compounded effects of brutal warfare, exposure to new bacteria and viruses (against which they had no immunity), and ultimately segregation on resource-starved reservations. The populations in Europe, Asia, and Africa had experienced regular gene flow during the 10,000-year period in which most kinds of livestock were being domesticated, giving them many generations of experience building up immunity against zoonotic diseases (those that can pass from animals to humans). In contrast, the residents of the Americas had been almost completely isolated during those millennia, so all these diseases swept through the Americas in rapid succession, creating a major loss of genetic diversity in the indigenous American population. It is estimated that between 50% and 95% of the indigenous American populations died during the first decades after European contact, around 500 years ago (Livi-Bacci 2006).
An urgent health challenge facing humans today involves human-induced population bottlenecks that produce antibiotic-resistant bacteria. Antibiotics are medicines prescribed to treat bacterial infections. The typical prescription includes enough medicine for ten days. People often feel better after less than ten days and sometimes decide to quit taking the medicine ahead of schedule. This is often a big mistake. The antibiotics have quickly killed off a large percentage of the bacteria—enough to reduce the symptoms and make you feel much better. However, this has created a bacterial population bottleneck. There are usually a small number of bacteria that survive those early days. If you take the medicine as prescribed for the full ten days, it’s quite likely that there will be no bacterial survivors. If you quit early, though, the survivors—who were the members of the original population who were most resistant to the antibiotic—will begin to reproduce again. Soon the infection will be back, possibly worse than before, and now all of the bacteria are resistant to the antibiotic that you had been prescribed.
Other activities that have contributed to the rise of antibiotic-resistant bacteria include the use of antibacterial cleaning products and the inappropriate use of antibiotics as a preventative measure in livestock or to treat infections that are viral instead of bacterial (viruses do not respond to antibiotics). In 2017, the World Health Organization published a list of twelve antibiotic-resistant pathogens that are considered top priority targets for the development of new antibiotics (World Health Organization 2017).
Founder Effects
Founder effects occur when members of a population leave the main or “parent” group and form a new population that no longer interbreeds with the other members of the original group. Similar to survivors of a population bottleneck, the newly founded population often has allele frequencies that are different from the original group. Alleles that may have been relatively rare in the parent population can end up being very common due to founder effect. Likewise, recessive traits that were seldom seen in the parent population may be seen frequently in the descendants of the offshoot population. One striking example of founder effect was first noted in the Dominican Republic in the 1970s. During a several-year period, eighteen children who had been born with female genitalia and raised as girls suddenly grew penises at puberty. This culture tended to value sons over daughters, so these transitions were generally celebrated. They labeled the condition guevedoces, which translates to “penis at twelve,” due to the average age at which this occurred. Scientists were fascinated by the phenomenon.
Genetic and hormonal studies revealed that the condition, scientifically termed , is an autosomal recessive syndrome that manifests when a child having both X and Y sex chromosomes inherits two nonfunctional (mutated) copies of the SRD5A2 gene (Imperato-McGinley and Zhu 2002). These children develop testes internally, but the 5-alpha reductase 2 steroid, which is necessary for development of male genitals in babies, is not produced. In absence of this male hormone, the baby develops female-looking genitalia (in humans, “female” is the default infant body form, if the full set of the necessary male hormones are not produced). At puberty, however, a different set of male hormones are produced by other fully functional genes. These hormones complete the male genital development that did not happen in infancy. This condition became quite common in the Dominican Republic during the 1970s due to founder effect—that is, the mutated SRD5A2 gene happened to be much more common among the Dominican Republic’s founding population than in the parent populations [the Dominican population derives from a mixture of indigenous Native American (Taino) peoples, West Africans, and Western Europeans]. Five-alpha reductase syndrome has since been observed in other small, isolated populations around the world.
Founder effect is closely linked to the concept of inbreeding, which in population genetics does not necessarily mean breeding with immediate family relatives. Instead, inbreeding refers to the selection of mates exclusively from within a small, closed population—that is, from a group with limited allelic variability. This can be observed in small, physically isolated populations but also can happen when cultural practices limit mates to a small group. As with founder effect, inbreeding increases the risk of inheriting two copies of any nonfunctional (mutant) alleles.
The Amish in the United States are a population that, due to their unique history and cultural practices, emerged from a small founding population and have tended to select mates from within their groups. The Old Order Amish population of Lancaster County, Pennsylvania, has approximately 50,000 current members, all of whom can trace their ancestry back to a group of approximately 80 individuals. This small founding population immigrated to the United States from Switzerland in the mid-1700s to escape religious persecution. Keeping to themselves, and selecting mates almost exclusively from their own communities, the Amish have become familiar with far more recessive traits than are seen in their parent population.

One of the genetic conditions that has been observed much more frequently in the Lancaster County Amish population is , which is an autosomal recessive disorder characterized by short stature (dwarfism), polydactyly [the development of more than five digits (fingers or toes) on the hands or feet], abnormal tooth development, and heart defects (see Figure 4.13). Among the general world population, Ellis-van Creveld syndrome is estimated to affect approximately 1 in 60,000 individuals; among the Old Order Amish of Lancaster County, the rate is estimated to be as high as 1 in every 200 births (D’Asdia et al. 2013). One of the great insights that has come from the study of founder effects is that a limited gene pool carries a much higher risk for genetic diseases. Genetic diversity in a population tends to greatly reduce these risks.
Gene Flow
The third force of evolution is traditionally called gene flow. As with genetic drift, this is a misnomer, because it refers to flowing alleles, not genes. (All populations of the same species share the same genes; it is the alleles of those genes that may vary.) Gene flow refers to the movement of alleles from one population to another. In most cases, gene flow can be considered synonymous with migration between populations.
Returning again to the example of our primordial cell population, let’s imagine that, after the volcanic fissure opened up in the ocean floor, wiping out the majority of the parent population, two surviving populations developed in the waters on opposite sides of the fissure. Ultimately, the lava from the fissure grew into a chain of islands that continued to provide a physical barrier between the populations, even after the lava had cooled.
In the initial generations after the eruption, due to founder effect, isolation, and random inheritance (genetic drift), the population to the west of the islands contained a vast majority of the ruffled membrane alleles while the eastern population predominantly carried the smooth alleles. Ocean currents in the area typically flowed from west to east, sometimes carrying cells (facilitating gene flow) from the western (ruffled) population to the eastern (smooth) population. Due to the ocean currents, it was almost impossible for any cells from the eastern population to be carried westward. Thus, for inheritance purposes, the western (ruffled) population remained isolated. In this case, the gene flow is uni-directional (going only in one direction) and unbalanced (only one population is receiving the new alleles).
Admixture
Among humans, gene flow is often described as admixture. In forensic cases, anthropologists and geneticists are often asked to estimate the ancestry of unidentified human remains to help determine whether they match any missing persons’ reports. This is one of the most complicated tasks in these professions because, while “race” or “ancestry” involves simple checkboxes on a missing person’s form, among humans today there are no truly distinct genetic populations. All modern humans are members of the same fully breeding-compatible species, and all human communities have experienced multiple episodes of gene flow (admixture), leading all humans today to be so genetically similar that we are all members of the same (and only surviving) human subspecies: Homo sapiens sapiens.
Gene flow between otherwise isolated non-human populations is often termed hybridization. One example of this involves the hybridization and spread of Africanized honey bees (a.k.a., “killer bees”) in the Americas. All honey bees worldwide are classified as Apis mellifera. Due to distinct adaptations to various environments around the world, there are 28 different subspecies of Apis mellifera.
During the 1950s, a Brazilian biologist named Warwick E. Kerr experimented with hybridizing African and European subspecies of honey bees to try to develop a strain that was better suited to tropical environments than the European honey bees that had long been kept by North American beekeepers. Dr. Kerr was careful to contain the reproductive queens and drones from the African subspecies, but in 1957, a visiting beekeeper accidentally released 26 queen bees of the subspecies Apis mellifera scutellate from southern Africa into the Brazilian countryside. The African bees quickly interbred with local European honey bee populations. The hybridized bees exhibited the more aggressively defensive behavior of the African strain, fatally or near-fatally attacking many humans and livestock that ventured too close to their hives. The Africanized bees spread throughout South America and reached Mexico and California by 1985. By 1990, permanent colonies had been established in Texas, and by 1997, 90% of trapped bee swarms around Tucson, Arizona, were found to be Africanized (Sanford 2006).
Another example involves the introduction of the Harlequin ladybeetle, Harmonia axyridis, native to East Asia, to other parts of the world as a “natural” form of pest control. Harlequin ladybeetles are natural predators of some of the aphids and other crop-pest insects. First introduced to North America in 1916, the “biocontrol” strains of Harlequin ladybeetles were considered to be quite successful in reducing crop pests and saving farmers substantial amounts of money. After many decades of successful use in North America, biocontrol strains of Harlequin ladybeetles were also developed in Europe and South America in the 1980s.
Over the seven decades of biocontrol use, the Harlequin ladybeetle had never shown any potential for development of wild colonies outside of its native habitat in China and Japan. New generations of beetles always had to be reared in the lab. That all changed in 1988, when a wild colony took root near New Orleans, Louisiana. Either through admixture with a native ladybeetle strain, or due to a spontaneous mutation, a new allele was clearly introduced into this population that suddenly enabled them to survive and reproduce in a wide range of environments. This population spread rapidly across the Americas and had reached Africa by 2004. In Europe, the invasive, North American strain of Harlequin ladybeetle admixed with the European strain (Figure 4.14), causing a population explosion (Lombaert et al. 2010). Even strains specifically developed to be flightless (to curtail the
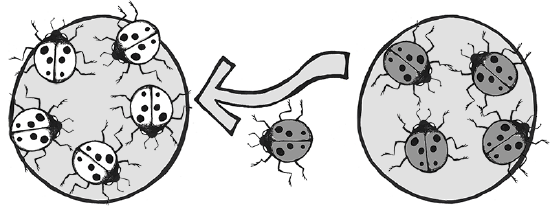
spreading) produced flighted offspring after admixture with members of the North American population (Facon et al. 2011). The fast-spreading, invasive strain has quickly become a disaster, out-competing native ladybeetle populations (some to the point of extinction), causing home infestations, decimating fruit crops, and contaminating many batches of wine with their bitter flavor after being inadvertently harvested with the grapes (Pickering et al. 2004).
Natural Selection
The final force of evolution is natural selection. This is the evolutionary process that Charles Darwin first brought to light, and it is what the general public typically evokes when considering the process of evolution. Natural selection occurs when certain phenotypes confer an advantage or disadvantage in survival and/or reproductive success. The alleles associated with those phenotypes will change in frequency over time due to this selective pressure. It’s also important to note that the advantageous allele may change over time (with environmental changes) and that an allele that had previously been benign may become advantageous or detrimental. Of course, dominant, recessive, and codominant traits will be selected upon a bit differently from one another. Because natural selection acts upon phenotypes rather than the alleles themselves, deleterious (disadvantageous) recessive alleles can be retained by heterozygotes without any negative effects.
In the case of our primordial ocean cells, up until now, the texture of their cell membranes has been benign. The frequencies of smooth to ruffled alleles, and smooth to ruffled phenotypes, has changed over time, due to genetic drift and gene flow. Let’s now imagine that the Earth’s climate has cooled to a point that the waters frequently become too cold for survival of the tiny bacteria that are the dietary staples of our smooth and ruffled cell populations. The way amoeba-like cells “eat” is to stretch out the cell membrane, almost like an arm, to encapsulate, then ingest, the tiny bacteria. When the temperatures plummet, the tiny bacteria populations plummet with them. Larger bacteria, however, are better able to withstand the temperature change.
The smooth cells were well-adapted to ingesting tiny bacteria but poorly suited to encapsulating the larger bacteria. The cells with the ruffled membranes, however, are easily able to extend their ruffles to encapsulate the larger bacteria. They also find themselves able to stretch their entire membrane to a much larger size than their smooth-surfaced neighbors, allowing them to ingest more bacteria at a given time and to go for longer periods between feedings. The smooth and ruffled traits, which had previously offered no advantage or disadvantage while food was plentiful, now are subject to natural selection. During the cold snaps, at least, the ruffled cells have a definite advantage. We can imagine that the western population that has mostly ruffled alleles will continue to do well, while the eastern population, which has a much smaller proportion of ruffled alleles, will gradually shift toward a higher frequency of ruffled alleles in future generations.
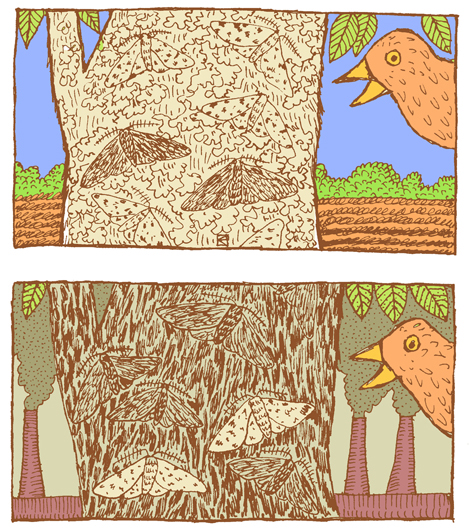
A classic example of natural selection involves the study of an insect called the peppered moth (Biston betularia) in England during the Industrial Revolution in the 1800s. Prior to the Industrial Revolution, the peppered moth population was predominantly light in color, with dark (pepper-like) speckles on the wings. The “peppered” coloration was very similar to the appearance of the bark and lichens that grew on the local trees (Figure 4.15). This helped to camouflage the moths as they rested on a tree, making it harder for moth-eating birds to find and snack on them. There was another phenotype that popped up occasionally in the population. These individuals were heterozygotes that carried an overactive, dominant pigment allele, producing a solid black coloration. As you can imagine, the black moths were much easier for birds to spot, making this phenotype a real disadvantage. The situation changed, however, as the Industrial Revolution took off. Large factories began spewing vast amounts of coal smoke into the air, blanketing the countryside, including the lichens and trees, in black soot. Suddenly, it was the light-colored moths that were easy for birds to spot and the black moths that held the advantage. The frequency of the dark pigment allele rose dramatically. By 1895, the black moth phenotype accounted for 98% of observed moths (Grant 1999).
Thanks to new environmental regulations in the 1960s, the air pollution in England began to taper off. As the soot levels decreased, returning the trees to their former, lighter color, this provided the perfect opportunity to study how the peppered moth population would respond. Repeated follow-up studies documented the gradual rise in the frequency of the lighter-colored phenotype. By 2003, the maximum frequency of the dark phenotype was 50% and in most parts of England had decreased to less than 10% (Cook 2003).
Directional, Balancing/Stabilizing, and Disruptive/Diversifying Selection
Natural selection can be classified as directional, balancing/stabilizing, or disruptive/diversifying, depending on how the pressure is applied to the population (Figure 4.16).
Both of the above examples of natural selection involve directional selection: the environmental pressures are favoring one phenotype over the other and causing the frequencies of the associated advantageous alleles (ruffled membranes, dark pigment) to
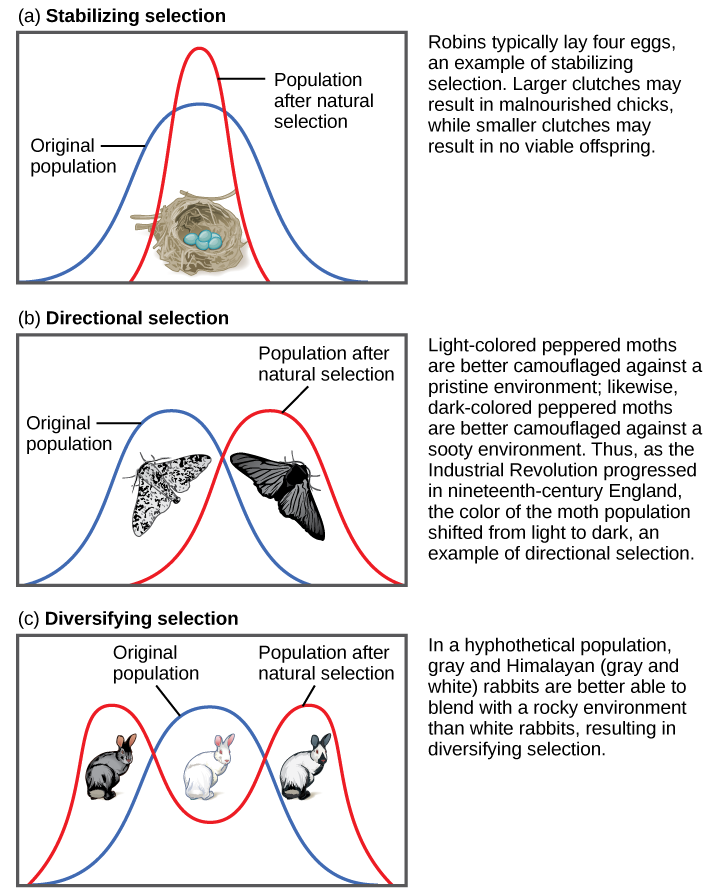
gradually increase. In the case of the peppered moths, the direction shifted three times: first, it was selecting for lighter pigment; then, with the increase in pollution, the pressure switched to selection for darker pigment; finally, with reduction of the pollution, the selection pressure shifted back again to favoring light-colored moths.
Balancing selection (a.k.a., stabilizing selection) occurs when selection works against the extremes of a trait and favors the intermediate phenotype. For example, humans maintain an average birth weight that balances the need for babies to be small enough not to cause complications during pregnancy and childbirth but big enough to maintain a safe body temperature after they are born. Another example of balancing selection is found in the genetic disorder called sickle cell anemia, which is featured in Case Study #2 (see below).
Disruptive selection (a.k.a., diversifying selection), the opposite of balancing selection, occurs when both extremes of a trait are advantageous. Since individuals with traits in the mid-range are selected against, disruptive selection can eventually lead to the population evolving into two separate species. Darwin believed that the many species of finches (small birds) found in the remote Galapagos Islands provided a clear example of disruptive selection leading to speciation. He observed that seed-eating finches either had large beaks, capable of eating very large seeds, or small beaks, capable of retrieving tiny seeds. The islands did not have many plants that produced medium-size seeds. Thus, birds with medium-size beaks would have trouble eating the very large seeds and would also have been inefficient at picking up the tiny seeds. Over time, Darwin surmised, this pressure against mid-size beaks may have led the population to divide into two separate species.
Case Study #2 : Sickle Cell Anemia
Sickle cell anemia is an autosomal recessive genetic disorder that affects millions of people worldwide. It is most common in Africa, countries around the Mediterranean Sea, and eastward as far as India. Populations in the Americas that have high percentages of ancestors from these regions also have high rates of sickle cell anemia. In the United States, it’s estimated that 72,000 people live with the disease, with one in approximately 1,200 Hispanic-American babies and one in every 500 African-American babies inheriting the condition (World Health Organization 1996).
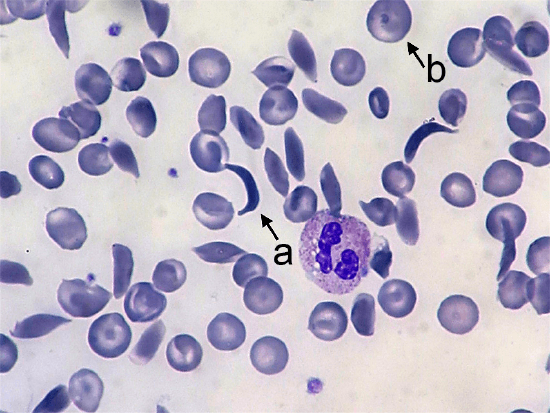
Sickle cell anemia affects the hemoglobin protein in red blood cells. Normal red blood cells are somewhat doughnut-shaped—round with a depression on both sides of the middle. They carry oxygen around the bloodstream to cells throughout the body. Red blood cells produced by the mutated form of the gene take on a stiff, sickle-like crescent shape when stressed by low oxygen or dehydration (Figure 4.17). Because of their elongated shape and the fact that they are stiff rather than flexible, they tend to form clumps in the blood vessels, inhibiting blood flow to adjacent areas of the body. This causes episodes of extreme pain and can cause serious problems in the oxygen-deprived tissues. The sickle cells also break down much more quickly than normal cells, often lasting only 20 days rather than the 120 days of normal cells. This causes an overall shortage of blood cells in the sickle cell patient, resulting in low iron (anemia) and problems associated with it such as extreme fatigue, shortness of breath, and hindrances to children’s growth and development.
The devastating effects of sickle cell anemia made its high frequency a pressing mystery. Why would an allele that is so deleterious in its homozygous form be maintained in a population at levels as high as the one in twelve African-Americans estimated to carry at least one copy of the allele? The answer turned out to be one of the most interesting cases of balancing selection in the history of genetic study.
While looking for an explanation, scientists noticed that the countries with high rates of sickle cell disease also shared a high risk for another disease called malaria, which is caused by infection of the blood by a Plasmodium parasite. These parasites are carried by mosquitoes and enter the human bloodstream via a mosquito bite. Once infected, the person will experience flu-like symptoms that, if untreated, can often lead to death. Researchers discovered that many people living in these regions seemed to have a natural resistance to malaria. Further study revealed that people who carry the sickle cell allele are far less likely to experience a severe case of malaria. This would not be enough of a benefit to make the allele advantageous for the sickle cell homozygotes who face shortened life spans due to sickle cell anemia. The real benefit of the sickle cell allele goes to the heterozygotes.
People who are heterozygous for sickle cell carry one normal allele, which produces the normal, round, red blood cells, and one sickle cell allele, which produces the sickle-shaped red blood cells. Thus, they have both the sickle and round blood cell types in their bloodstream. They produce enough of the round red blood cells to avoid the symptoms of sickle cell anemia, but they have enough sickle cells to provide protection from malaria.
When the Plasmodium parasites infect an individual, they begin to multiply in the liver, but then must infect the red blood cells to complete their reproductive cycle. When the parasites enter sickle-type cells, the cells respond by taking on the sickle shape. This prevents the parasite from circulating through the bloodstream and completing its life cycle, greatly inhibiting the severity of the infection in the sickle cell heterozygotes compared to non-sickle-cell homozygotes. See chapter 14 for more discussion of sickle cell anemia.
Sexual Selection
Sexual selection is an aspect of natural selection in which the selective pressure specifically affects reproductive success (the ability to successfully breed and raise offspring) rather than survival. Sexual selection favors traits that will attract a mate. Sometimes these sexually appealing traits even carry greater risks in terms of survival.
A classic example of sexual selection involves the brightly colored feathers of the peacock. The peacock is the male sex of the peafowl genera Pavo and Afropavo. During mating season, peacocks will fan their colorful tails wide and strut in front of the peahens in a grand display. The peahens will carefully observe these displays and will elect to mate with the male that they find the most appealing. Many studies have found that peahens prefer the males with the fullest, most colorful tails. While these large, showy tails provide a reproductive advantage, they can be a real burden in terms of escaping predators. The bright colors and patterns as well as the large size of the peacock tail make it difficult to hide. Once predators spot them, peacocks also struggle to fly away, with the heavy tail trailing behind and weighing them down (Figure 4.18). Some researchers have argued that the increased risk is part of the appeal for the peahens: only an especially strong, alert, and healthy peacock would be able to avoid predators while sporting such a spectacular tail.
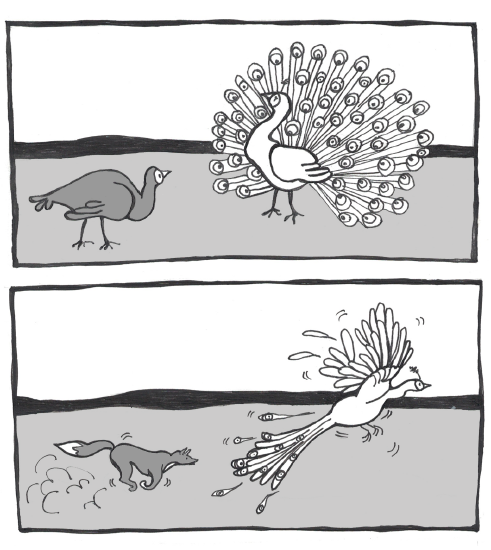 Figure \(\PageIndex{13}\): Showy peacock tail advantages (impressing peahens) and disadvantages (becoming easier prey).
Figure \(\PageIndex{13}\): Showy peacock tail advantages (impressing peahens) and disadvantages (becoming easier prey).It’s important to keep in mind that sexual selection relies on the trait being present throughout mating years. Reflecting back on Case Study #1, the examination of the NF1 genetic disorder, some might find it surprising that half of the babies born with NF1 inherited it from a parent. Given how disfiguring the symptoms can become, and the fact that the disorder is autosomal dominant and fully penetrant (meaning it has no unaffected carriers), it may seem surprising that sexual selection doesn’t exert more pressure against the mutated alleles. One important factor is that, while the neurofibromas typically begin to appear during puberty, they usually emerge only a few at a time and may grow very slowly. Many NF1 patients don’t experience the more severe or disfiguring symptoms until later in life, long after they have started families of their own.
Some researchers prefer to classify sexual selection separately, as a fifth force of evolution. The traits that underpin mate selection are entirely natural, of course. Research has shown that subtle traits, such as the type of pheromones (hormonal odors related to immune system alleles) someone emits and how those are perceived by the immune system genotype of the “sniffer,” may play crucial and subconscious roles in whether we find someone attractive or not (Chaix et al. 2008).
SPECIAL TOPIC: THE REAL PRIMORDIAL CELLS—DICTYOSTELIUM DISCOIDEUM
The amoeba-like primordial cells that were used as recurring examples throughout this chapter are inspired by actual research that is truly fascinating. In 2015, Gareth Bloomfield and colleagues reported on their genomic study of the social amoeba Dictyostelium discoideum (a.k.a., “slime molds,” although technically they are amoebae, not molds). Strains of these amoebae have been grown in research laboratories for many decades and are useful in studying phagocytosis and micropinocytosis—the mechanisms that amoeboid single-celled organisms use to ingest food and liquid. For simplification of our examples in this chapter, our amoeba-like cells remained ocean dwellers. Wild Dictyostelium discoideum, however, live in soil and feed on soil bacteria by growing ruffles in their membranes that reach out to encapsulate the bacterial cell. Laboratory strains, however, are typically raised on liquid media (agar) in Petri dishes, which is not suitable for the wild-type amoebae. It was widely known that the laboratory strains must have developed mutations in one or more genes to allow them to ingest the larger nutrient particles in the agar and larger volumes of liquid, but the genes involved were not known.
Bloomfield and colleagues performed genomic testing on both the wild and the laboratory strains of Dictyostelium discoideum. Their discovery was astounding: every one of the laboratory strains carried a mutation in the NF1 gene, the very same gene associated with Neurofibromatosis Type 1 (NF1) in humans. The antiquity of this massive, easily mutated gene is incredible. It originated in a common ancestor to humans and these amoebae and has been retained in both lineages ever since. As seen in Dictyostelium discoideum, breaking the gene can be advantageous. Without a functioning copy of the neurofibromin protein, the cell membrane is able to form much larger feeding structures, allowing the NF1 mutants to ingest larger particles and larger volumes of liquid. For these amoebae, this may provide dietary flexibility that functions somewhat like an insurance policy for times when the food supply is limited.
Dictyostelium discoideum are also interesting in that they typically reproduce asexually, but under certain conditions, one cell will convert into a “giant” cell, which encapsulates surrounding cells, transforming into one of three sexes. This cell will undergo meiosis, producing gametes that must combine with one of the other two sexes in order to produce viable offspring. This ability for sexual reproduction may be what allows Dictyostelium discoideum to benefit from the advantages of NF1 mutation, while also being able to restore the wild type NF1 gene in future generations.
What does this mean for humans living with NF1? Well, understanding the role of the neurofibromin protein in the membranes of simple organisms like Dictyostelium discoideum may help us to better understand how it functions and malfunctions in the sheaths of human neurons. It’s also possible that the mutability of the NF1 gene confers certain advantages to humans as well. Alleles of the NF1 gene have been found to reduce one’s risk for alcoholism (Repunte-Canonigo et al. 2015), opiate addiction (Sanna et al. 2002), Type 2 diabetes (Martins et al. 2016), and hypomusicality (a lower-than-average musical aptitude; Cota et al. 2018). This research is ongoing and will be exciting to follow in the coming years.

